
Notre objectif est de répondre précisément à vos besoins, en ajoutant une touche originale à vos projets, tout en respectant rigoureusement les délais et les coûts. Ensemble, construisons notre chemin vers le succès.
+55
ans de savoir-faire
+500
clients
Explorez comment personnaliser ces éléments pour une gestion plus efficace de vos équipements.
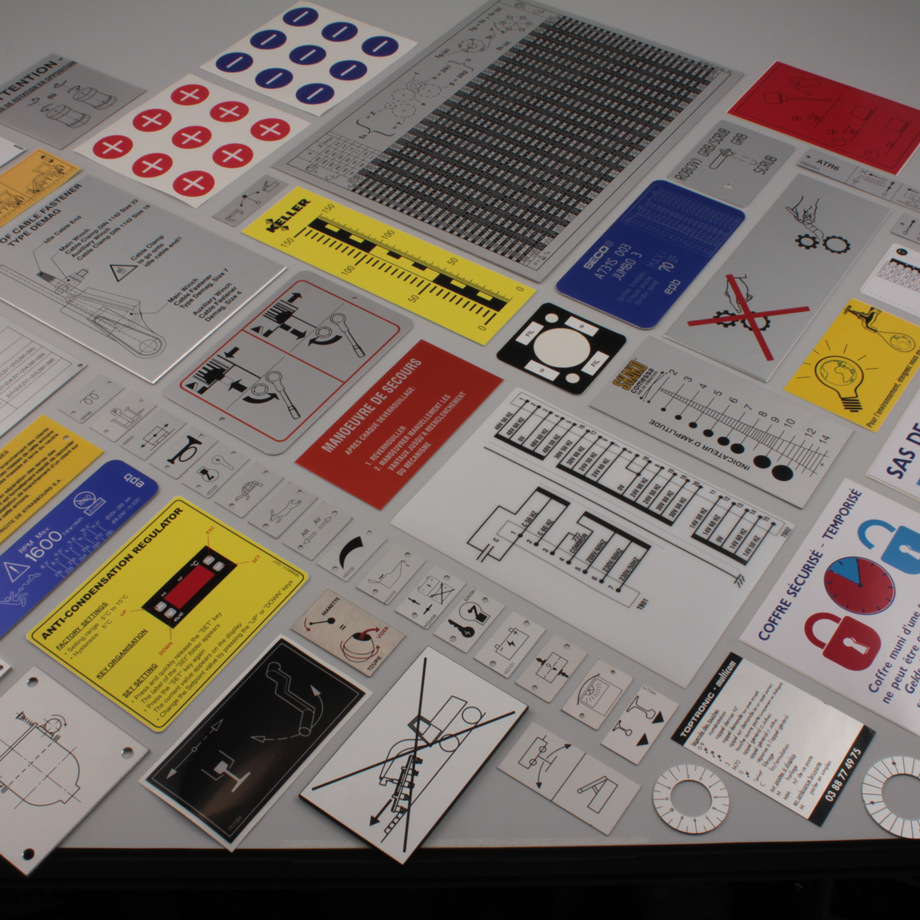
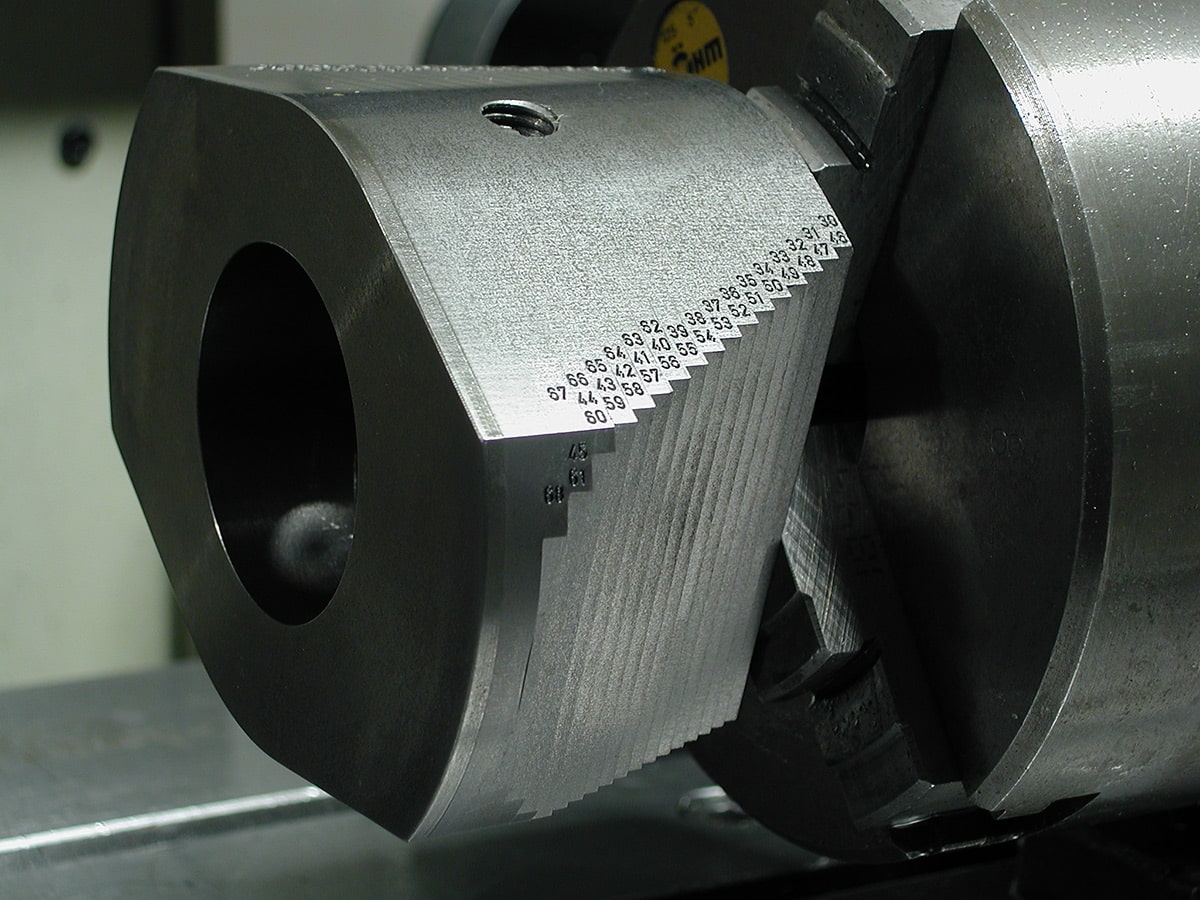
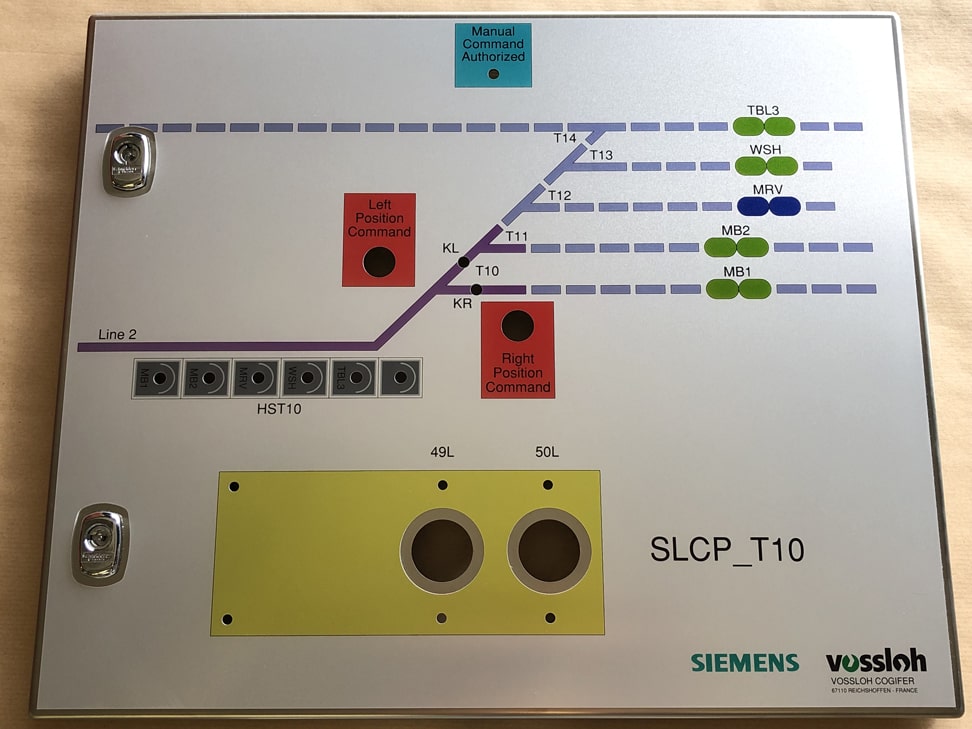



Aménagez votre espace de travail avec du mobilier technique sur mesure
Mettez en avant votre entreprise avec une signalétique impactante adaptée à vos besoins.


Qualité
Environnement
Fiabilité
Innovation
Découvrez les différentes techniques à notre disposition pour transformer vos idées en réalité. Qu’il s’agisse d’impression, de découpe, de gravure, d’usinage ou de conception en 3D, nous sommes le partenaire idéal pour vous accompagner dans tous vos projets.

Bureau d'étude
Studio Graphique
Gravure et usinage

Sérigraphie et tampographie

Marquage, découpe laser

Impression numérique

Découpe numérique

Façonnage - assemblage

Nouveau site internet
Dans le cadre de la digitalisation de notre entreprise, nous avons engagé la refonte c... [...]
Kellersign
Faites nous part de votre projet de marquage, création, prototypage, etc... une demande de devis simple, rapide et sans engagement.
Prenons contact